User Guide
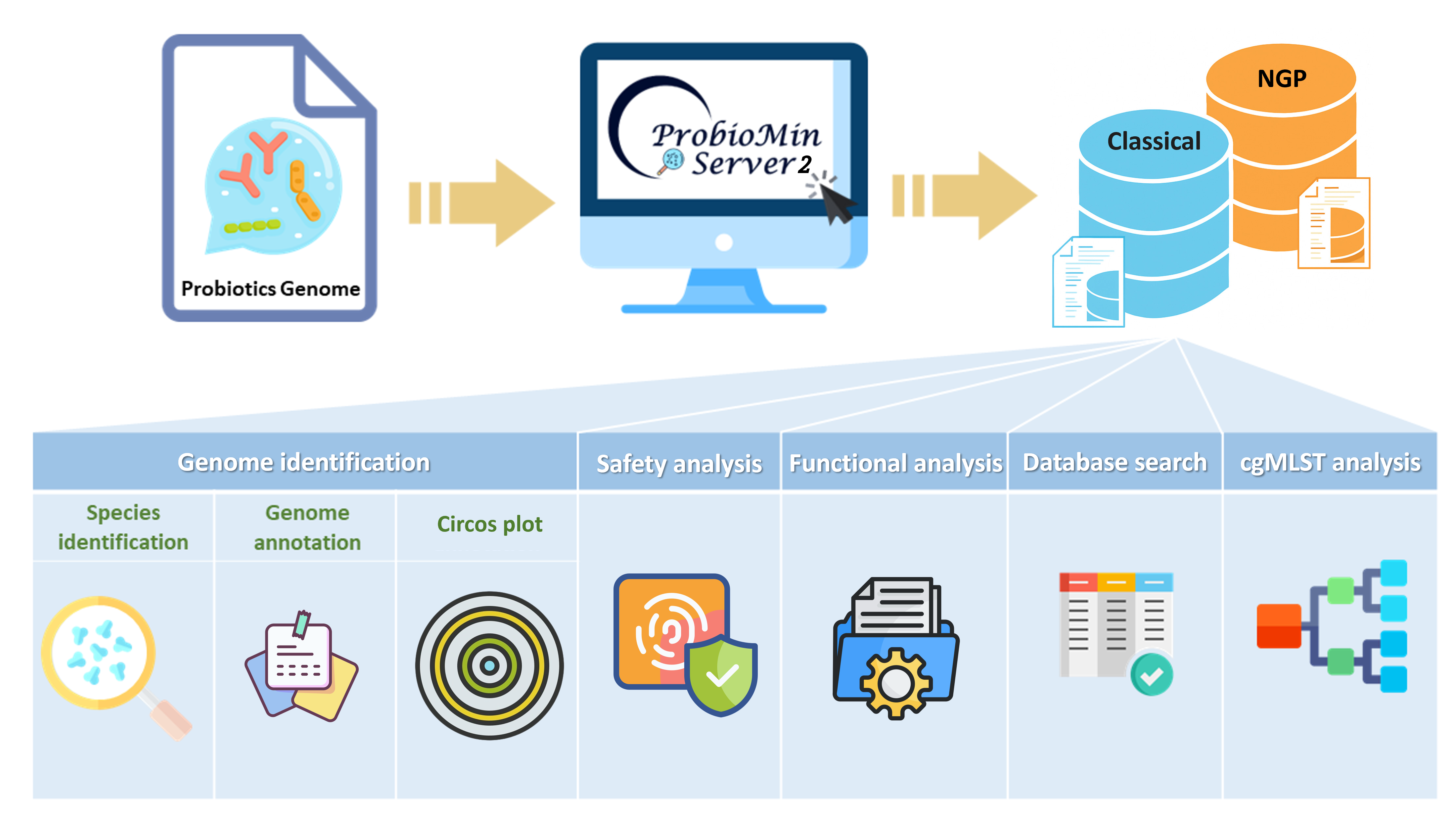
Graphical Abstract
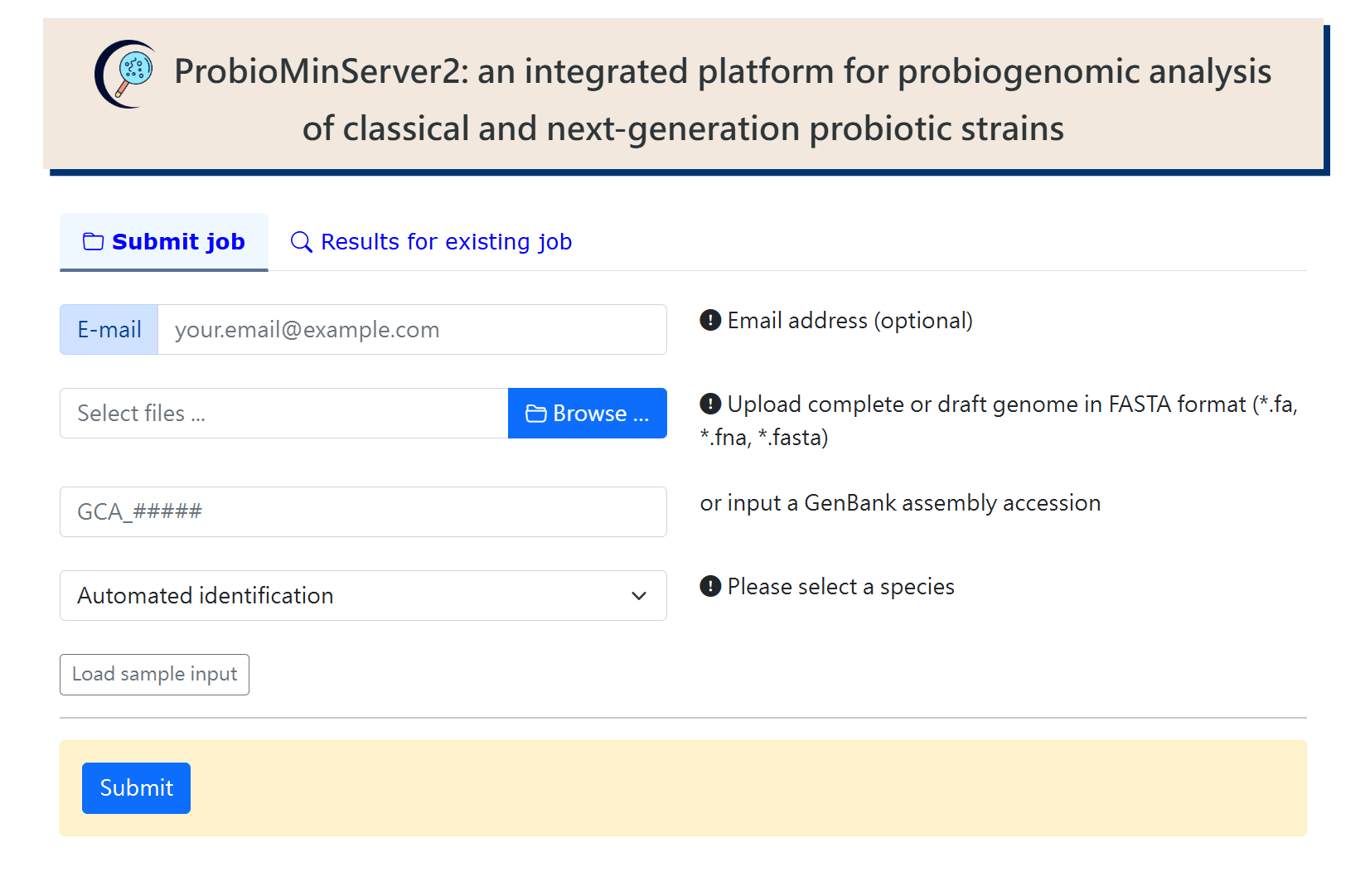
(I)ANALYSIS
Upload complete or draft genome file
Search results for an existing job
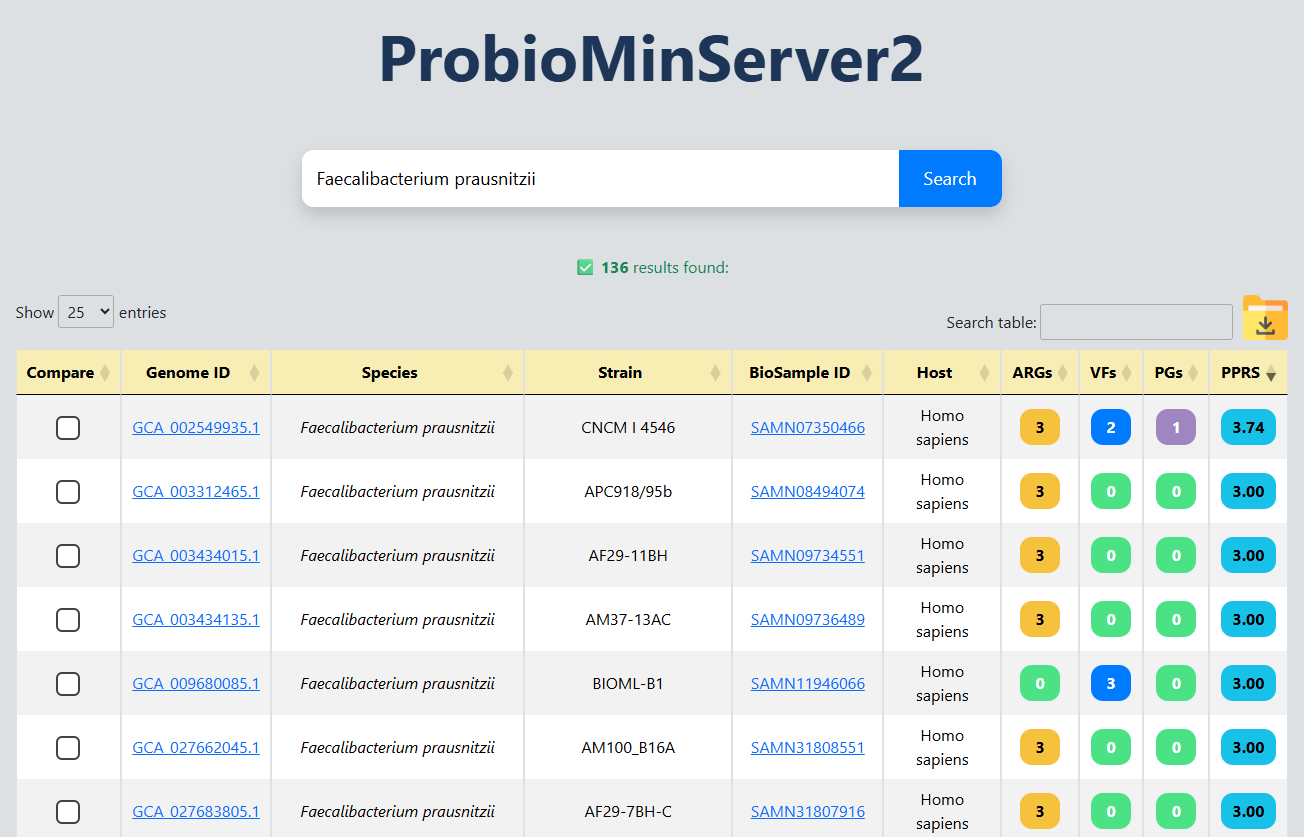
(II)BROWSE
Search and browse database

Detail of a species
(III)RESULTS
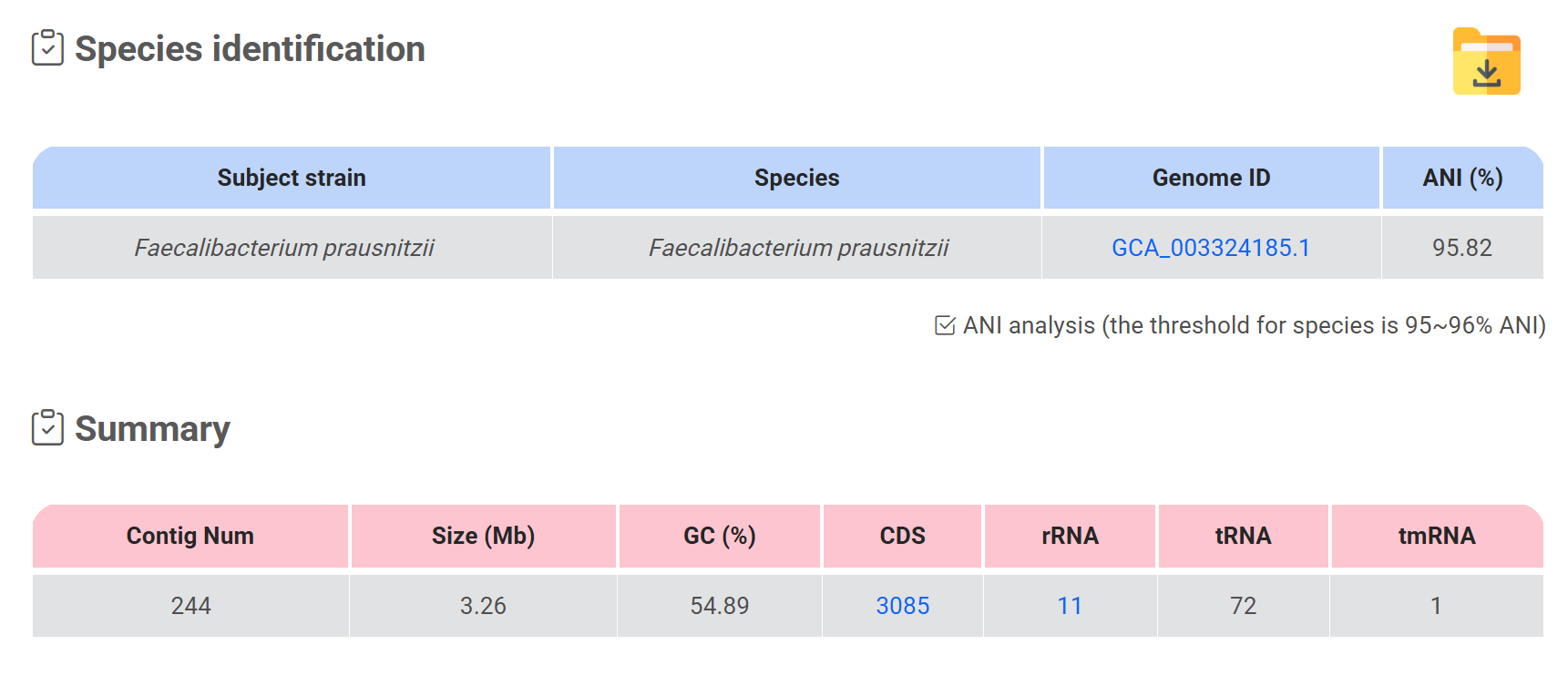
Species identification & Summary
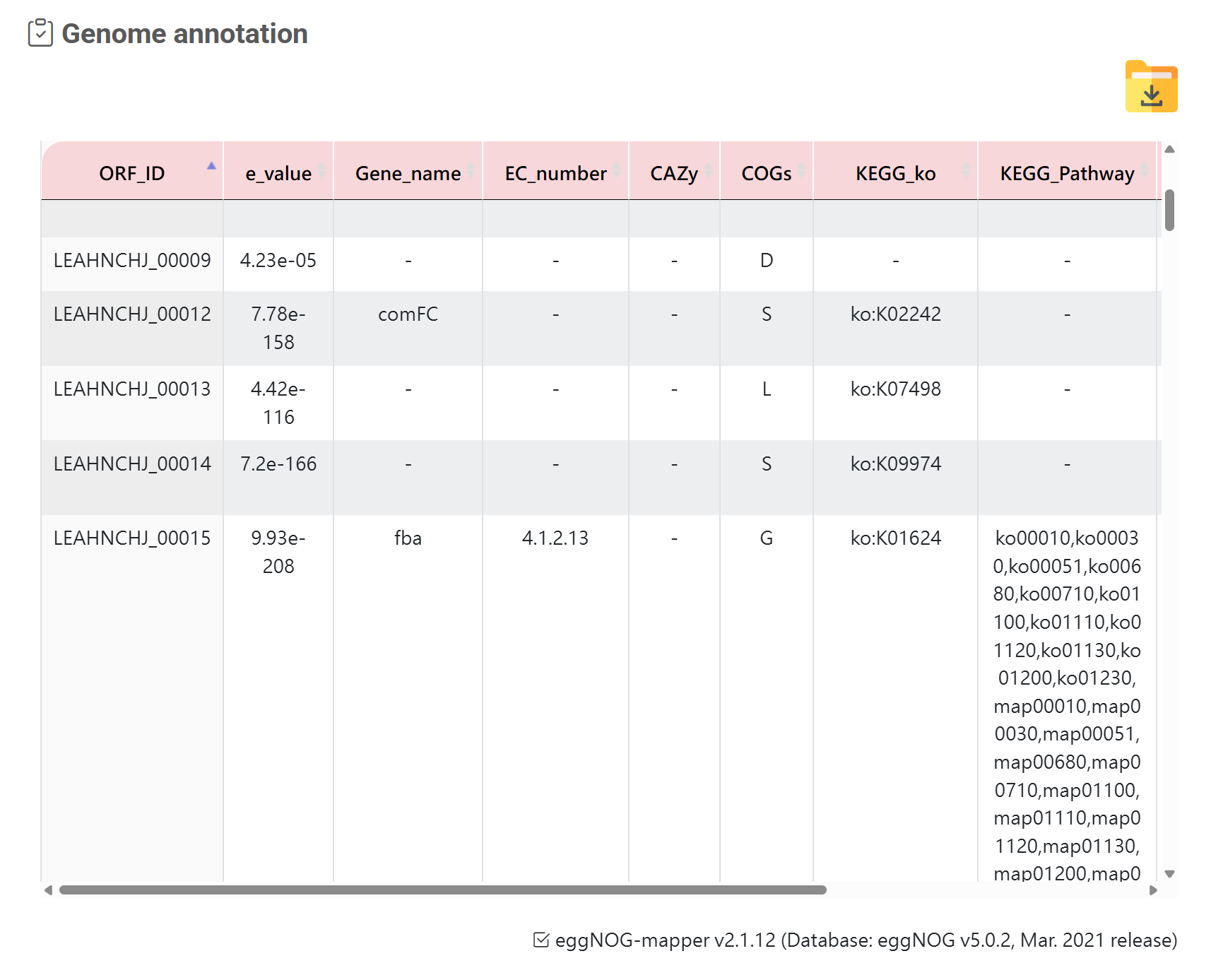
Annotation
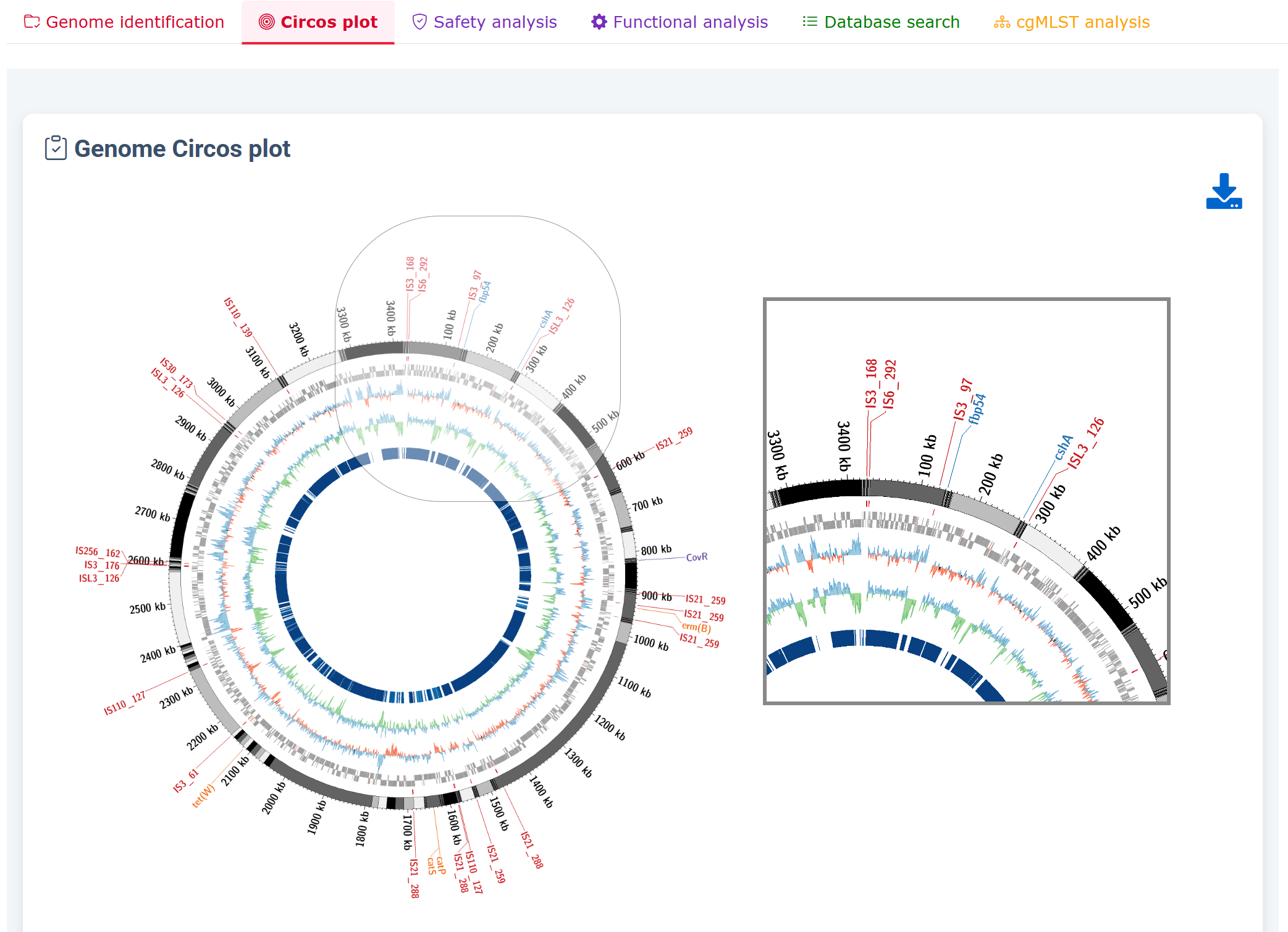
Circos plot
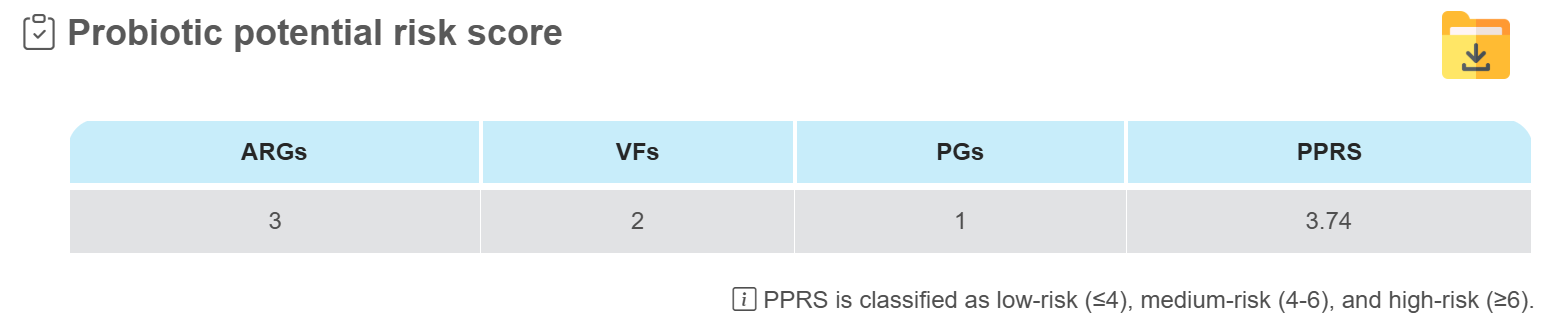
Probiotic potential risk score
Antimicrobial resistance gene
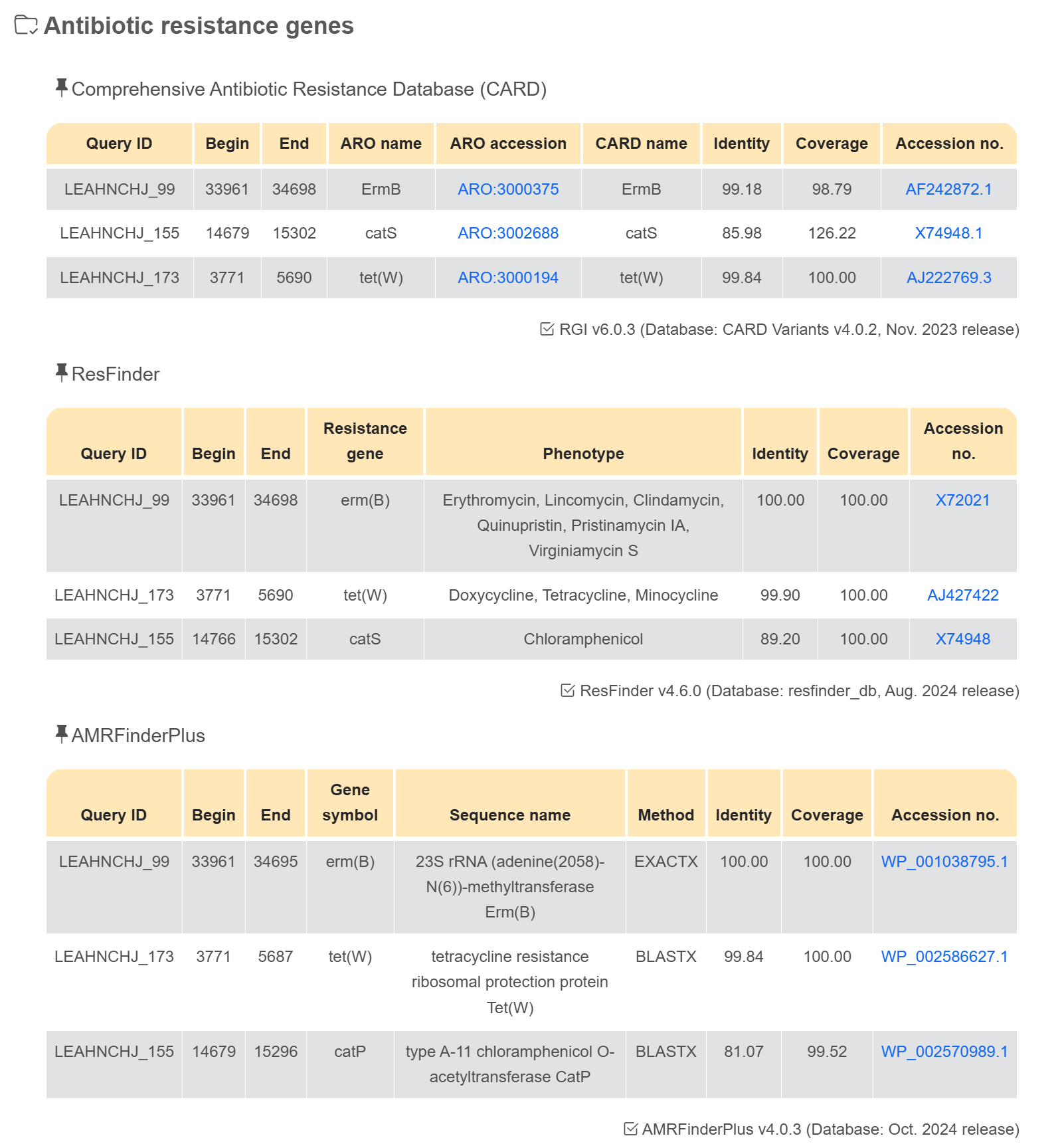
Virulence factor
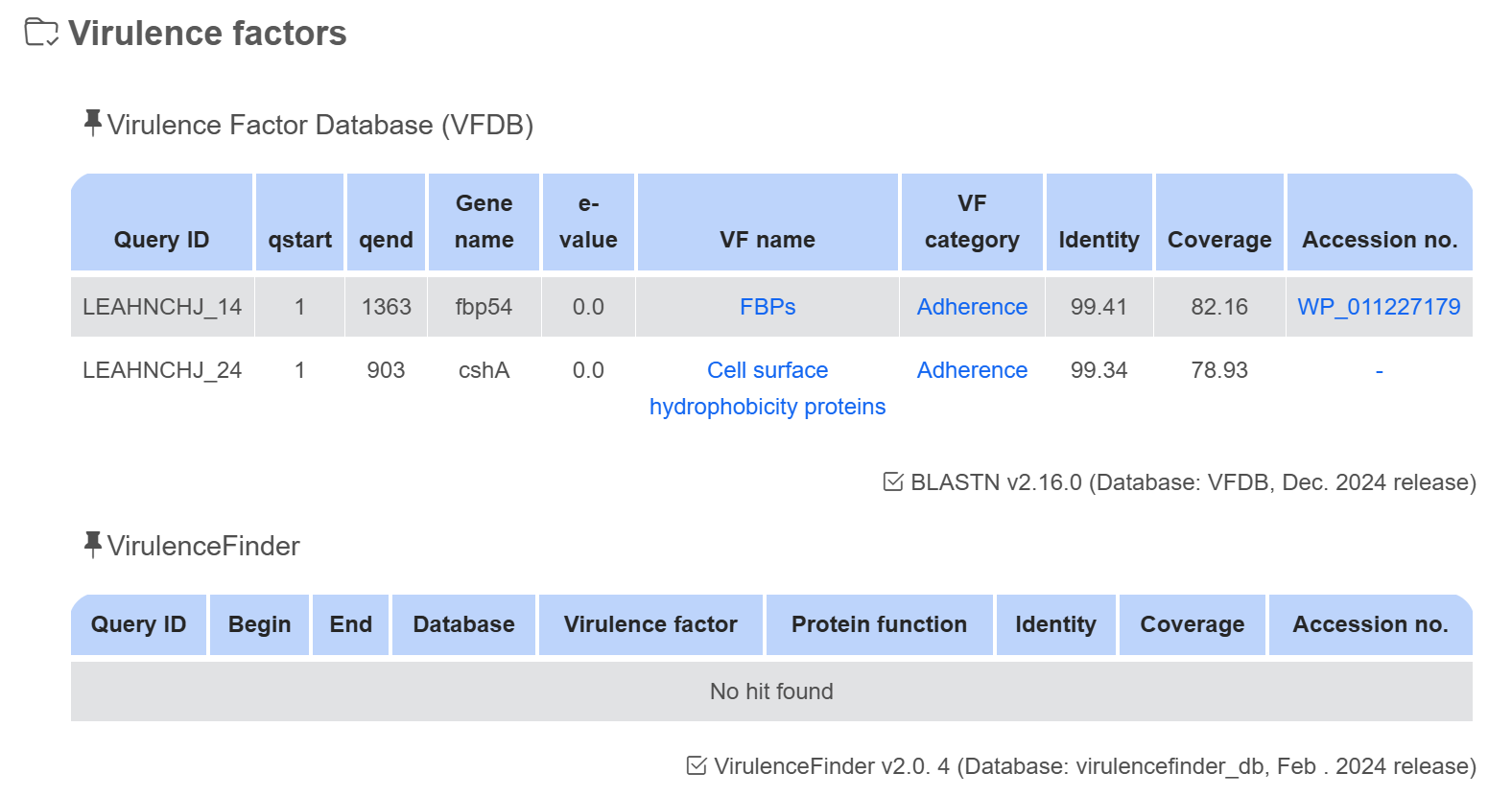
Pathogenic genes
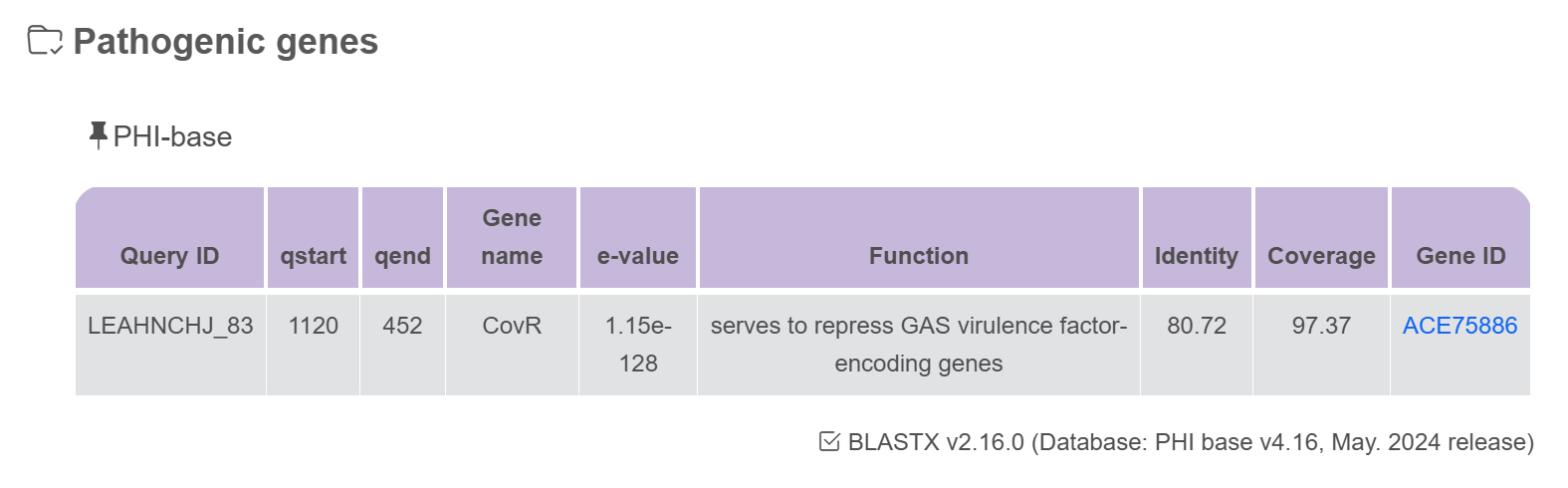
Plasmid
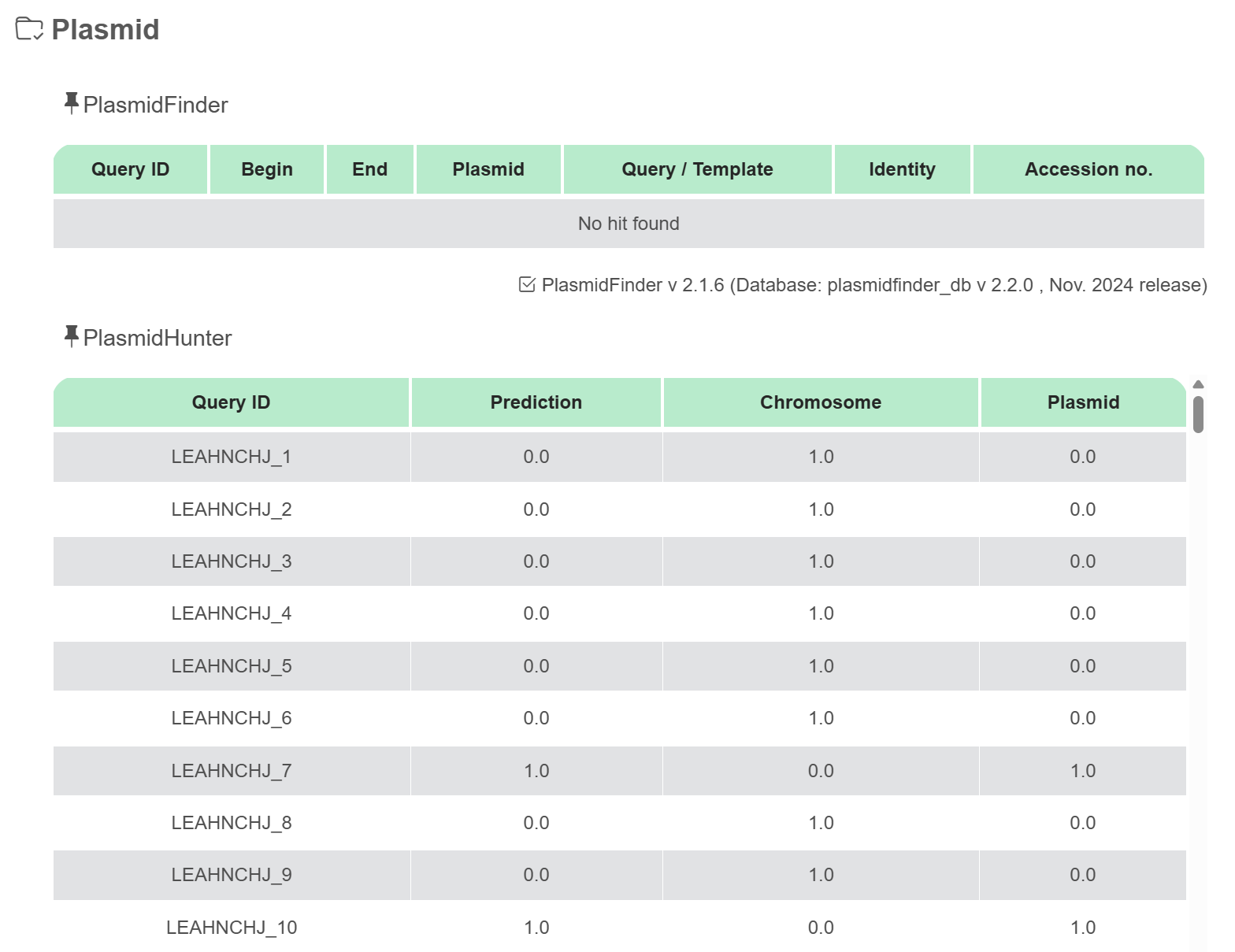
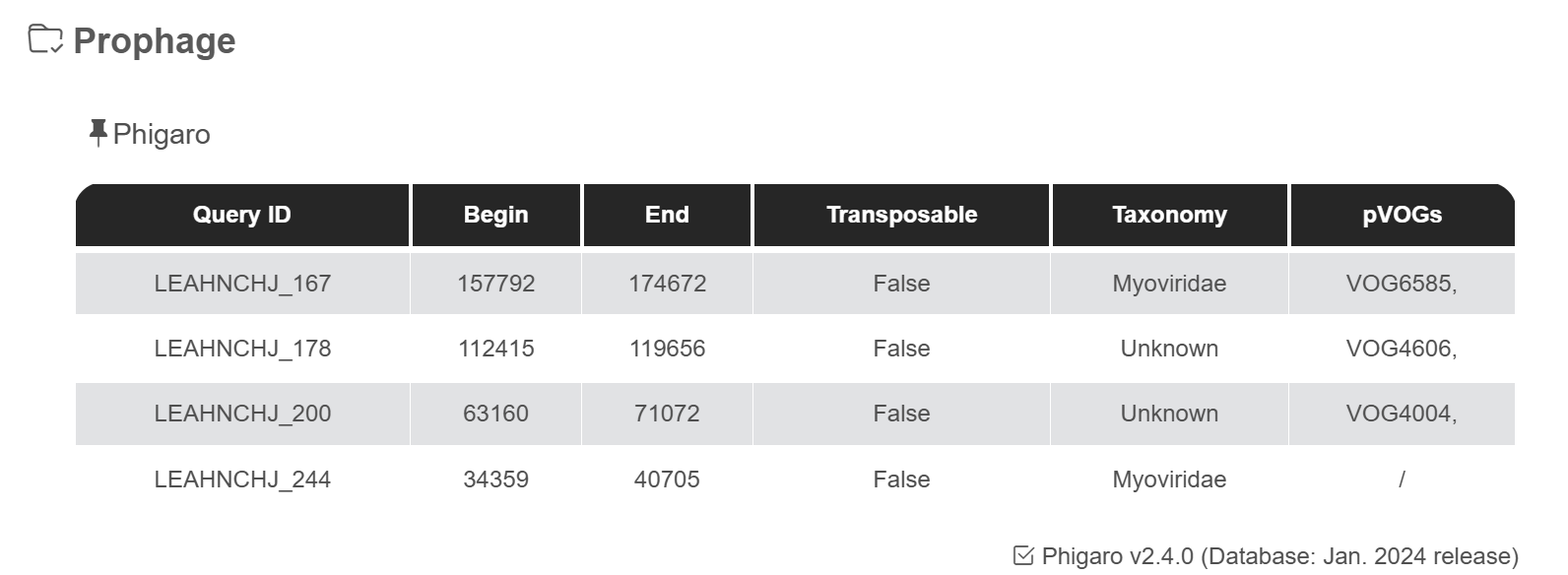
Phigaro
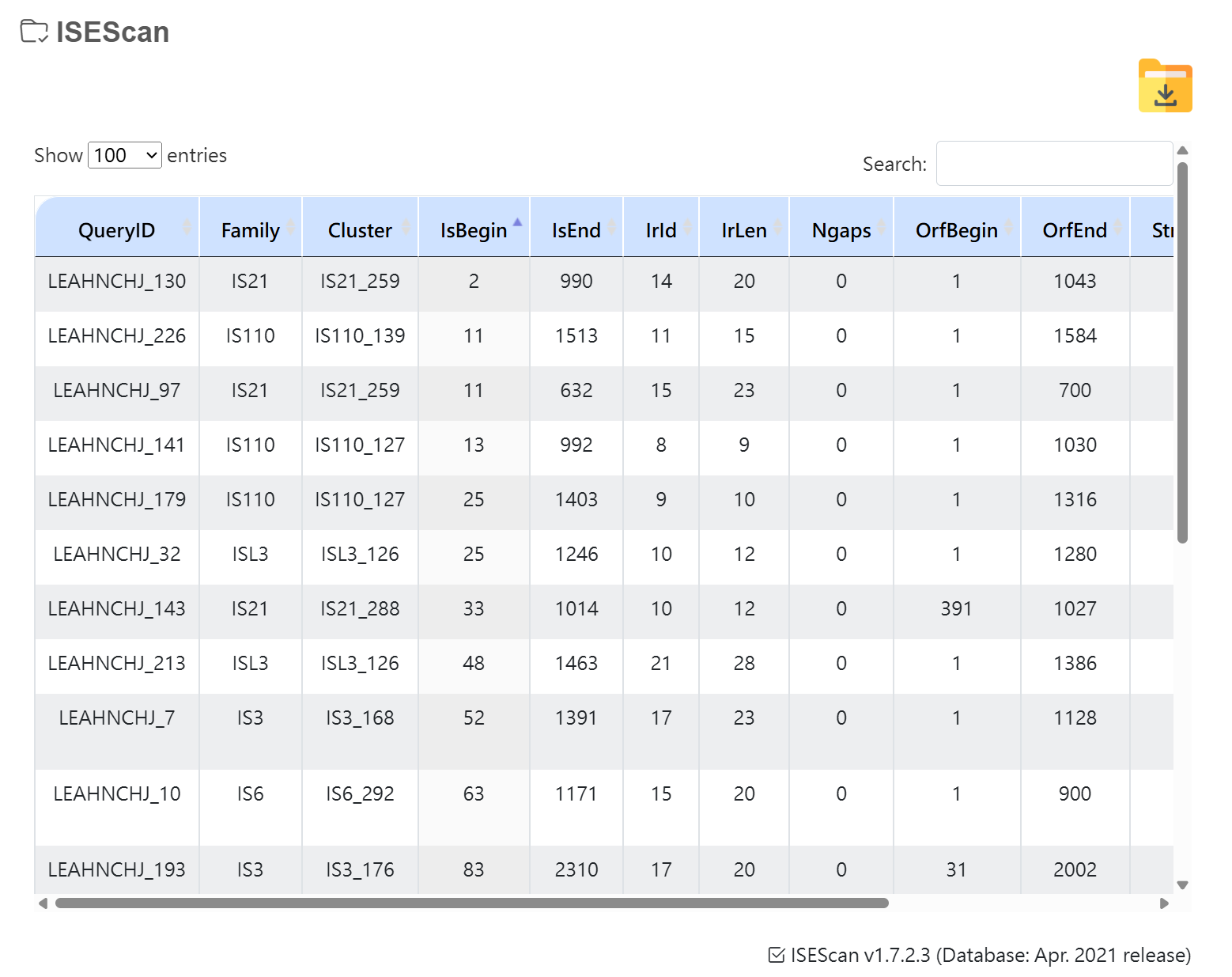
ISEScan
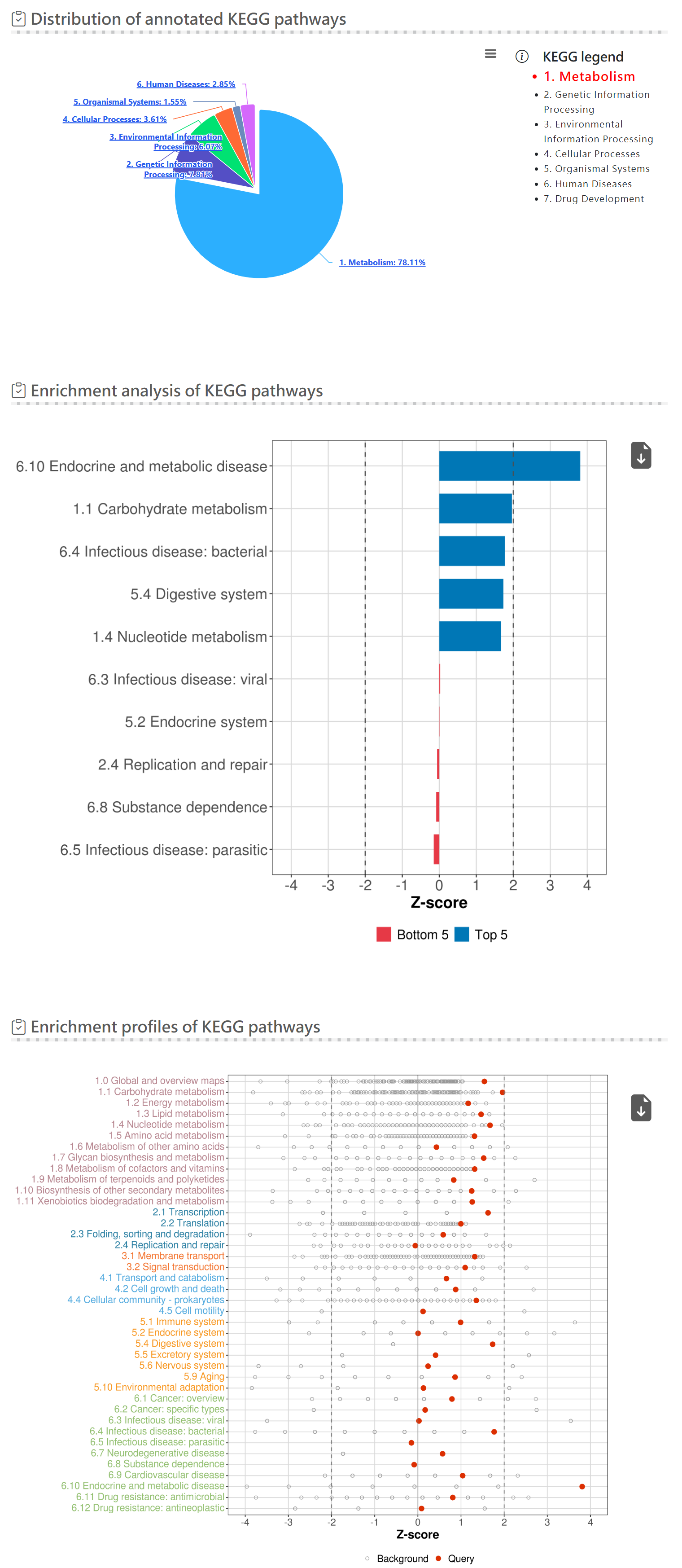
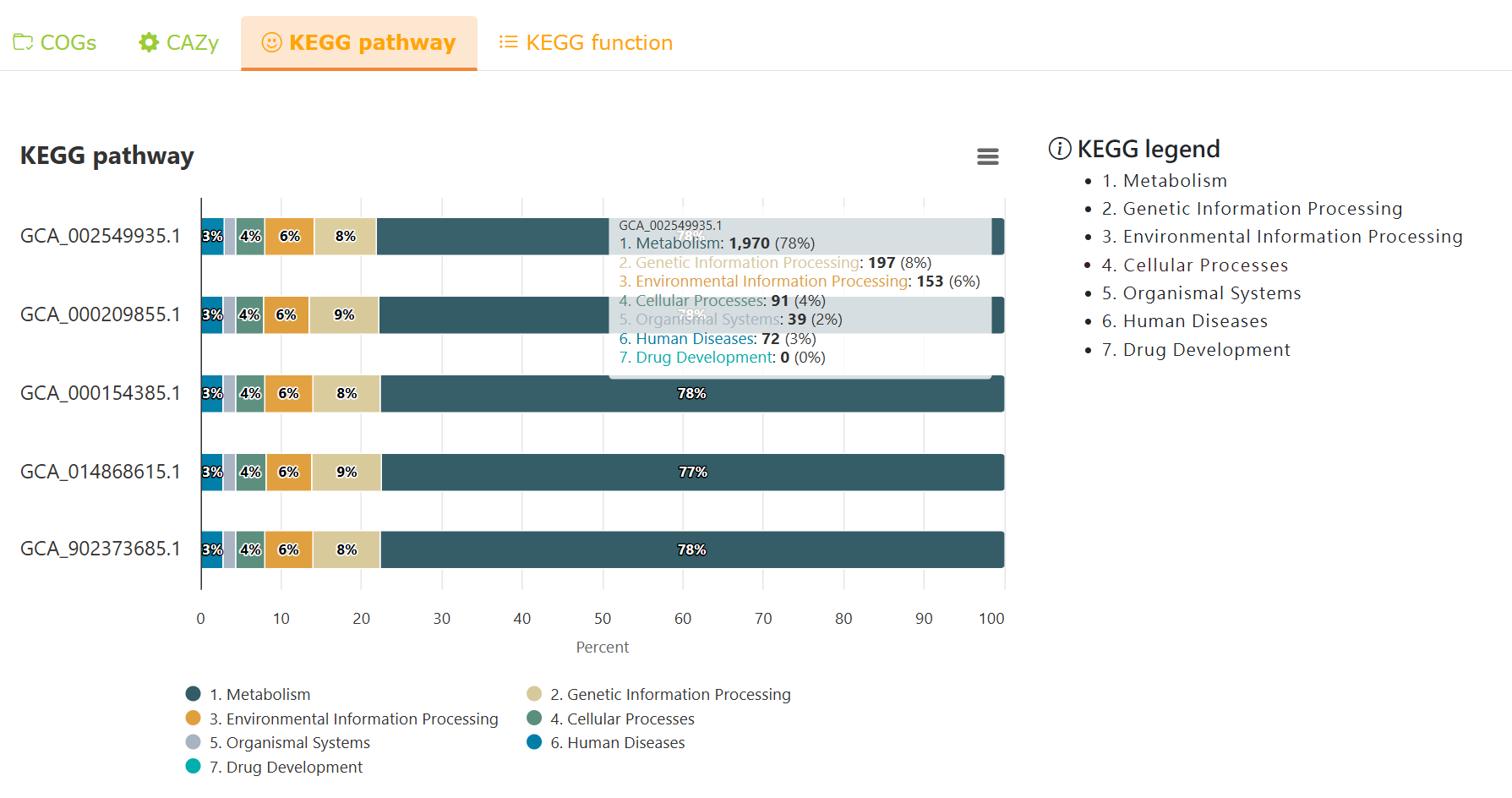
Functional analysis - KEGG pathway
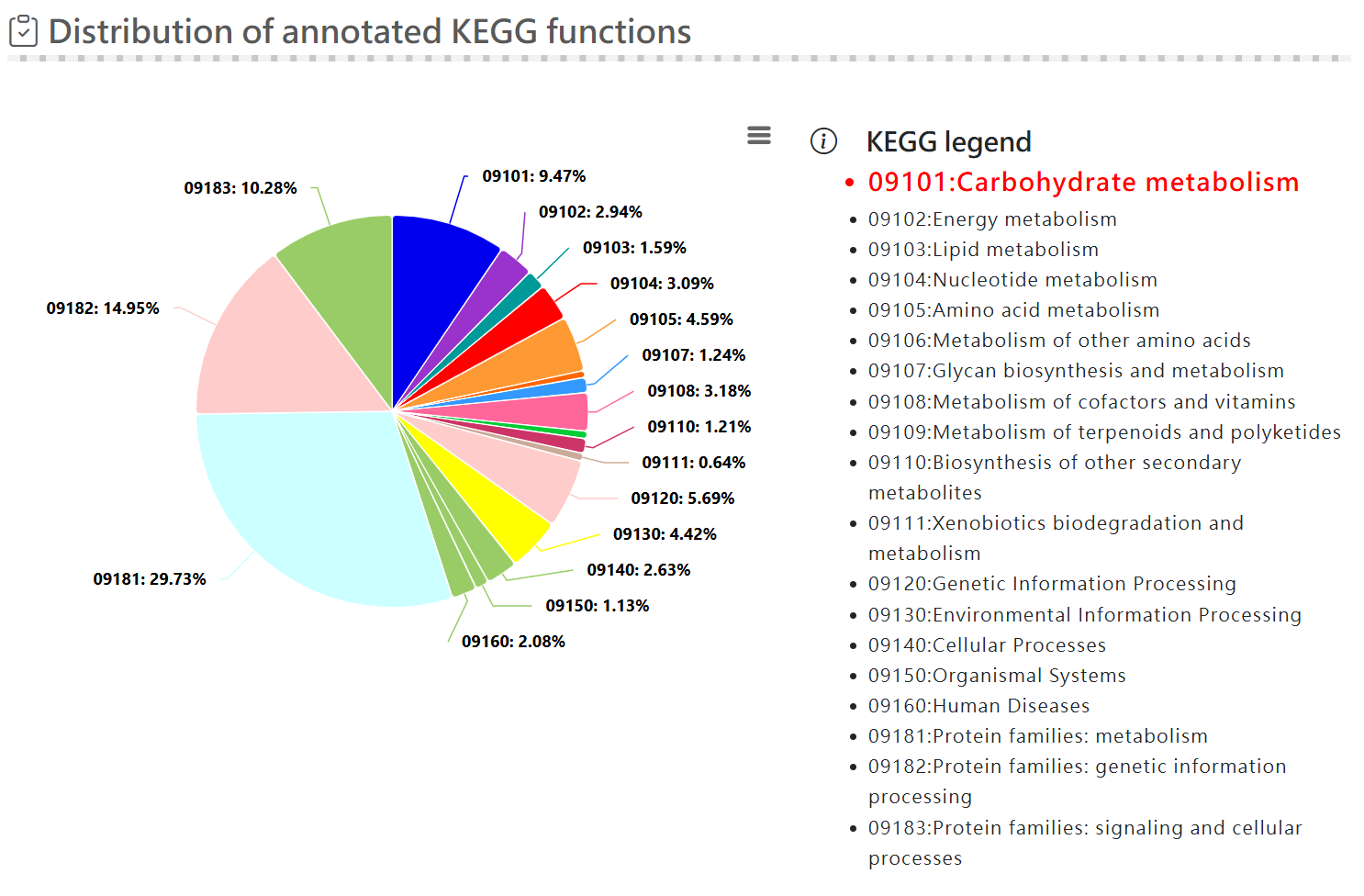
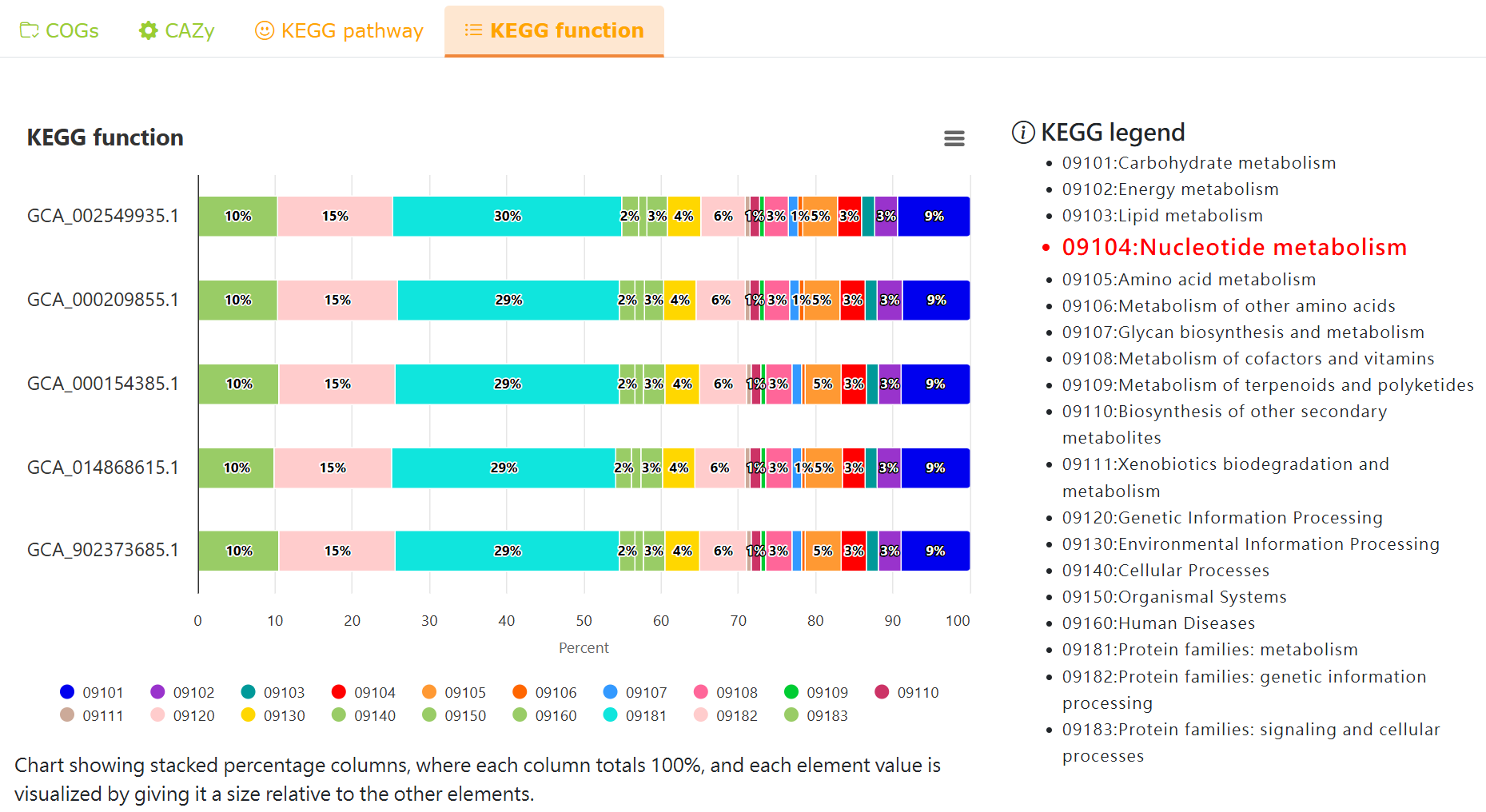
Functional analysis - KEGG function
Functional analysis - COGs
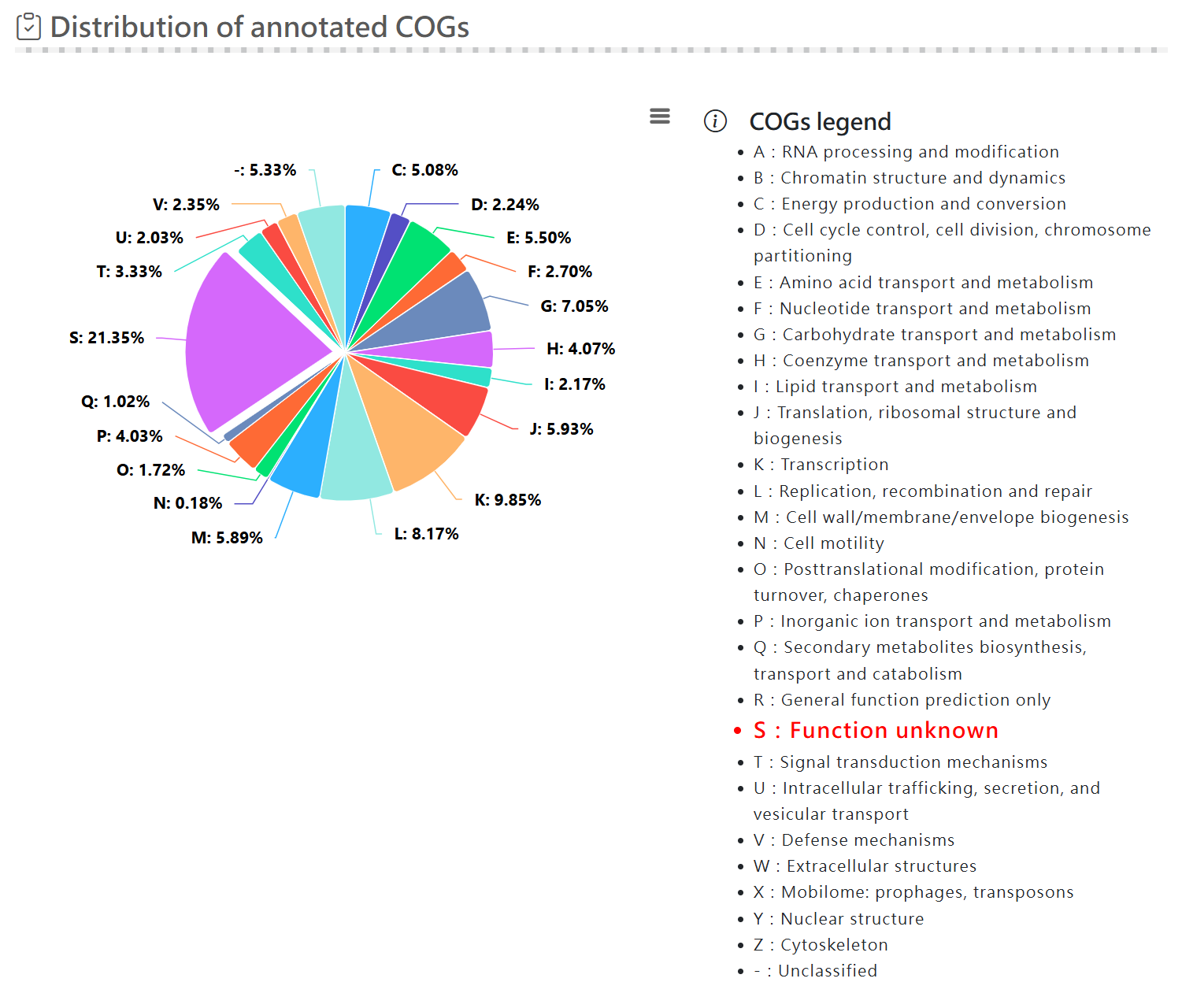
Functional analysis - CAZy
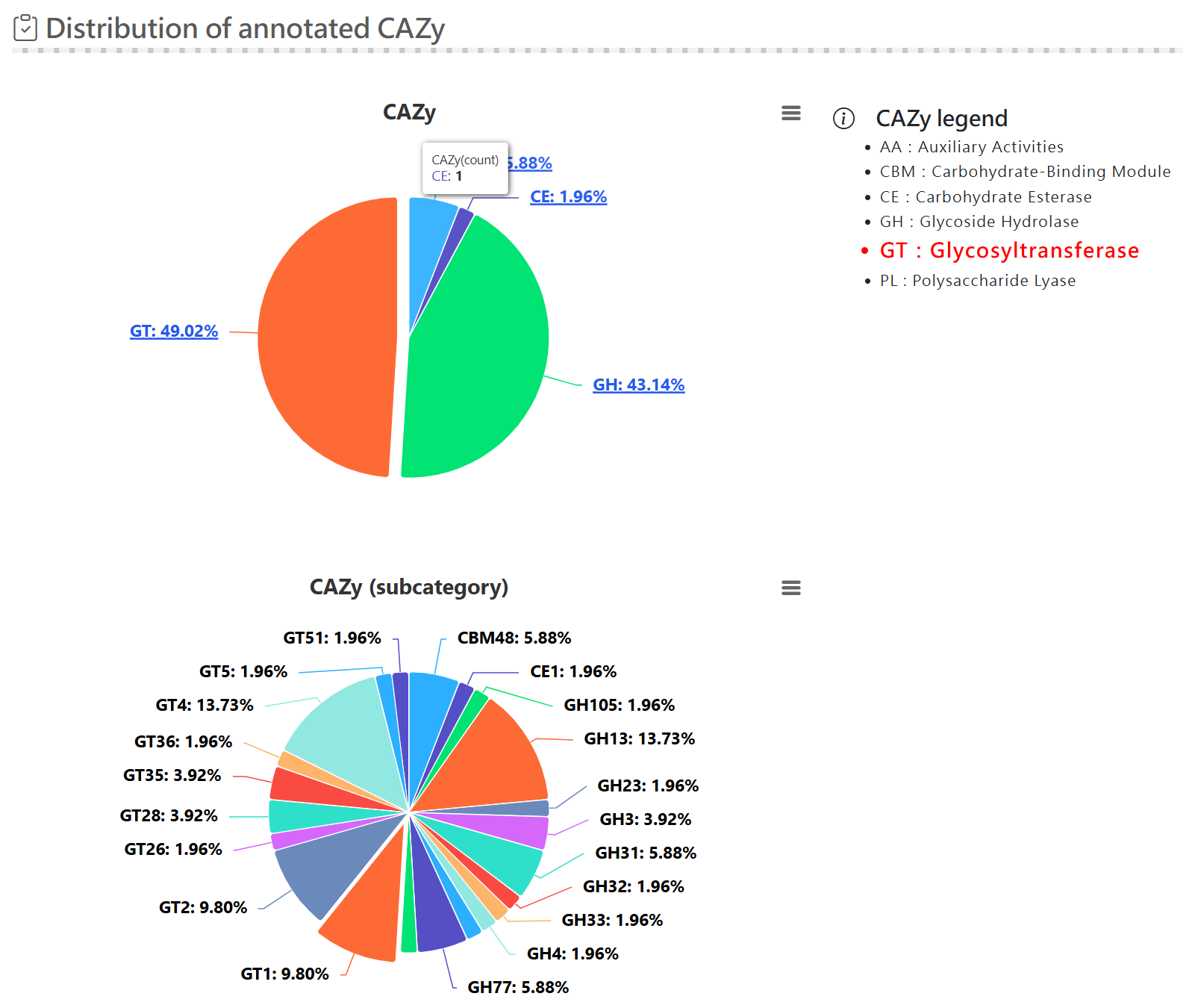
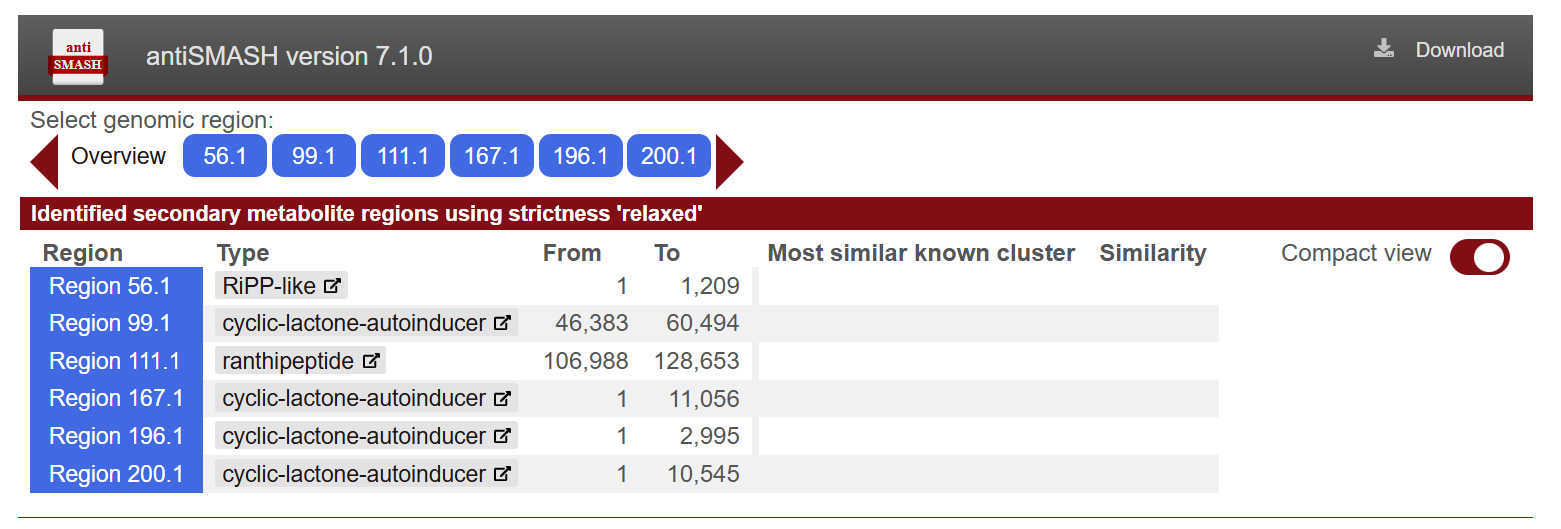
Functional analysis - antiSMASH
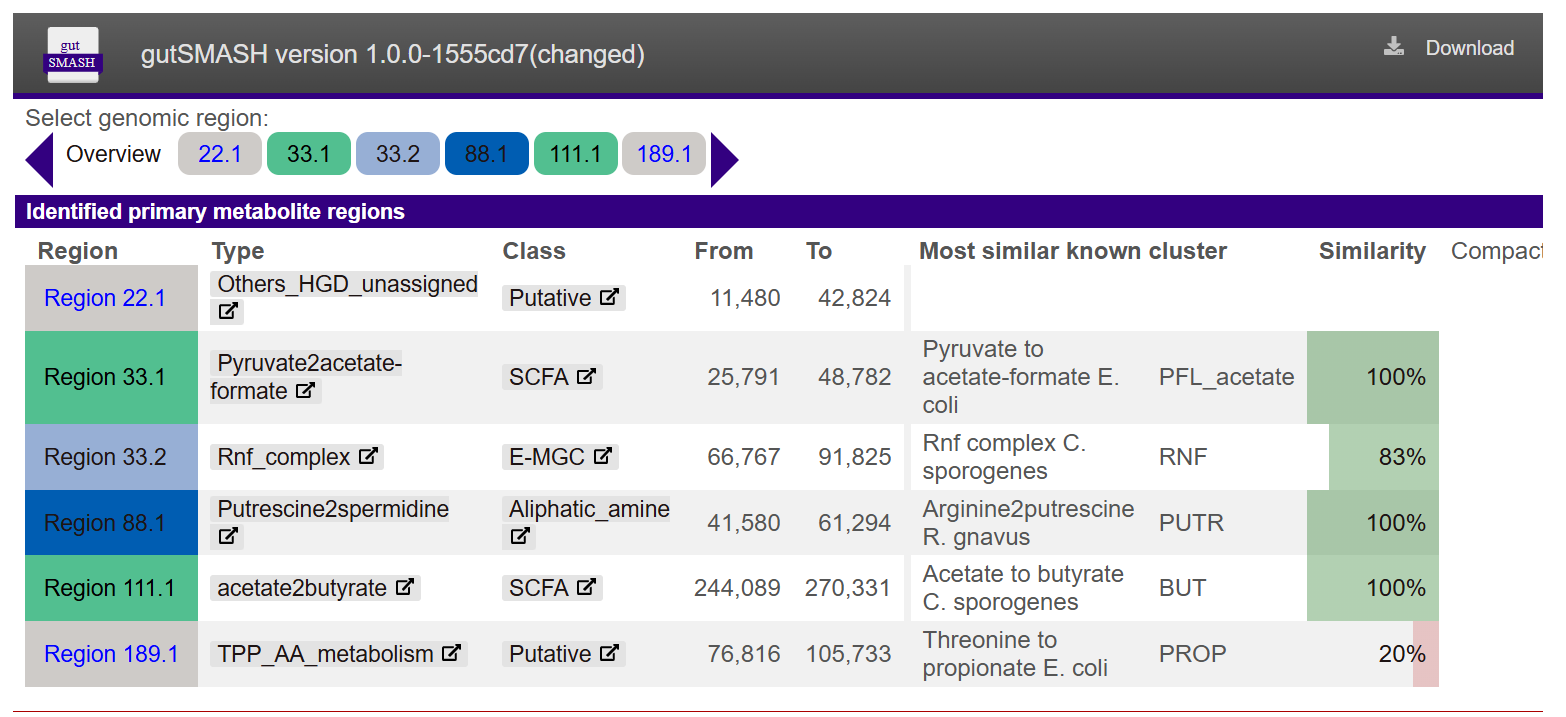
Functional analysis - gutSMASH
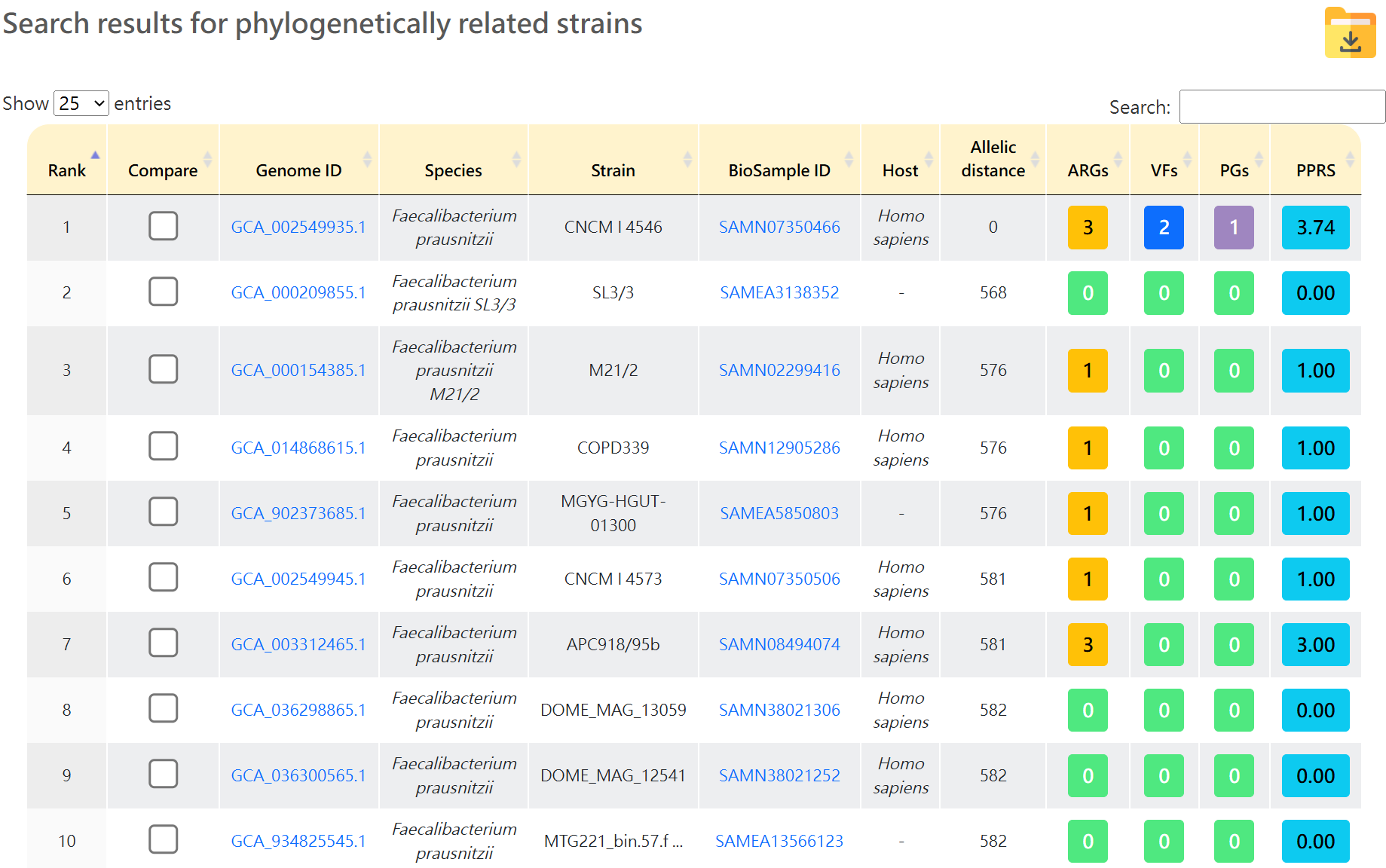
Search results for phylogenetically related strains
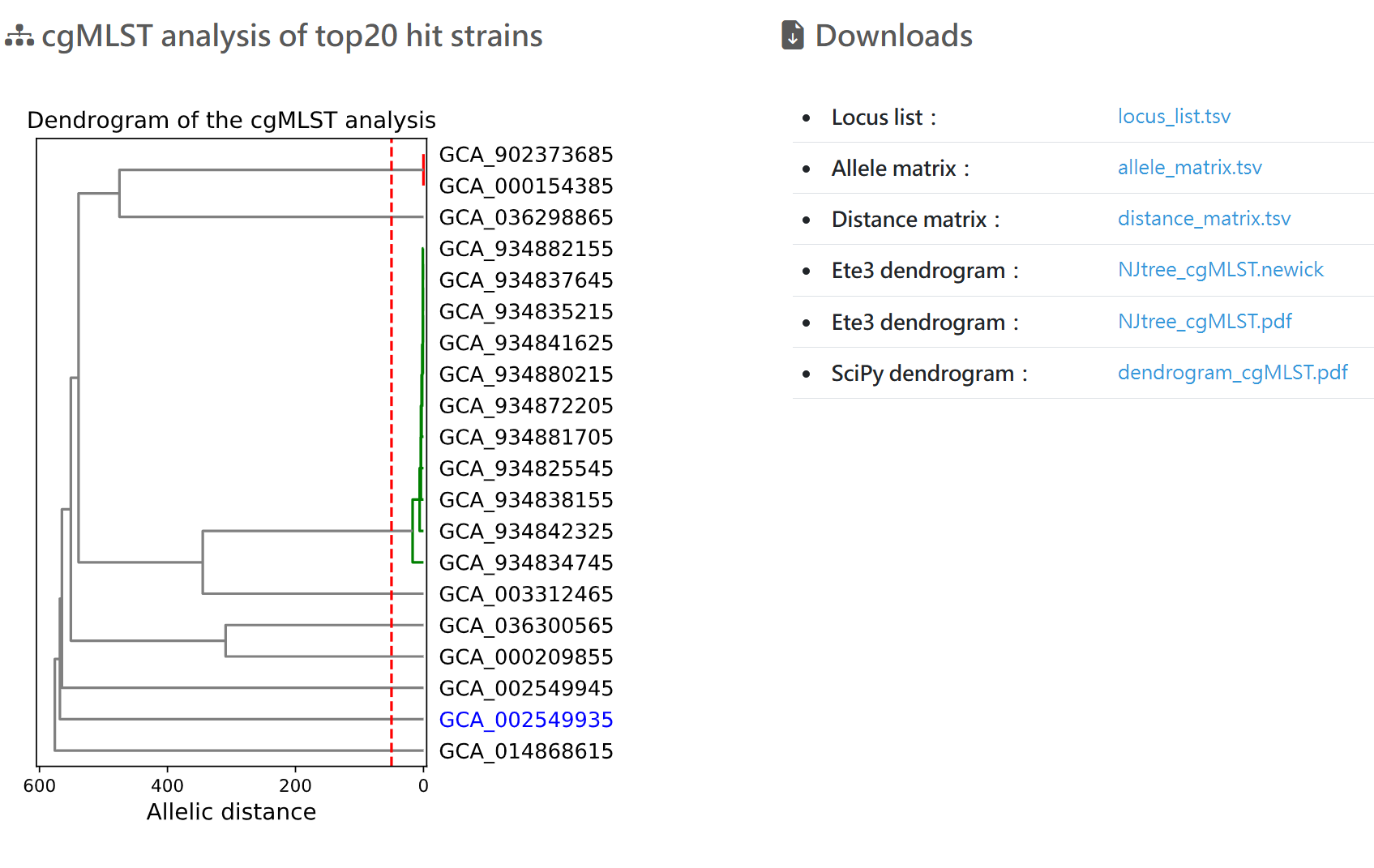
cgMLST analysis of top20 hit strains
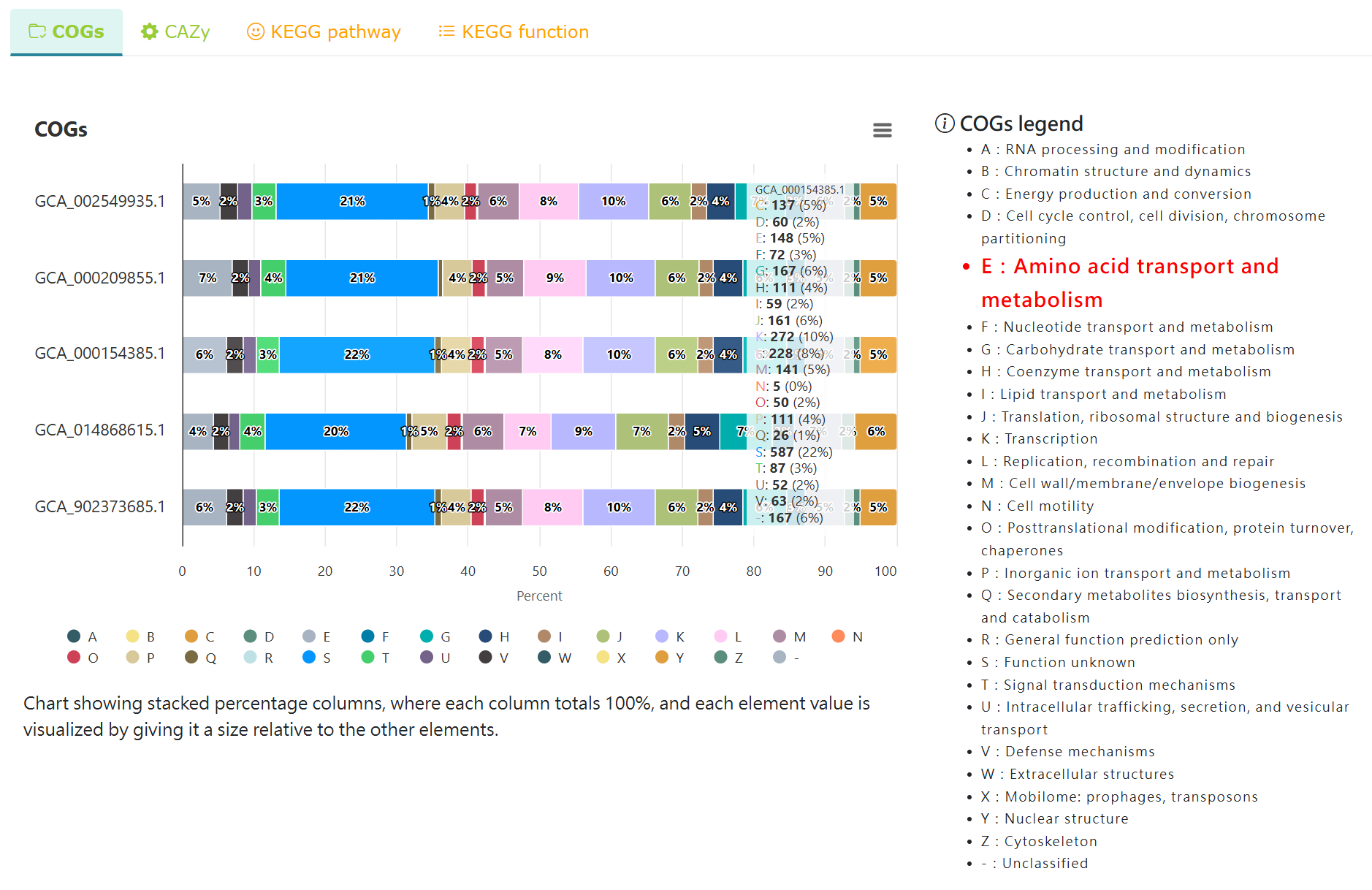
Comparative COGs analysis of the selected strains
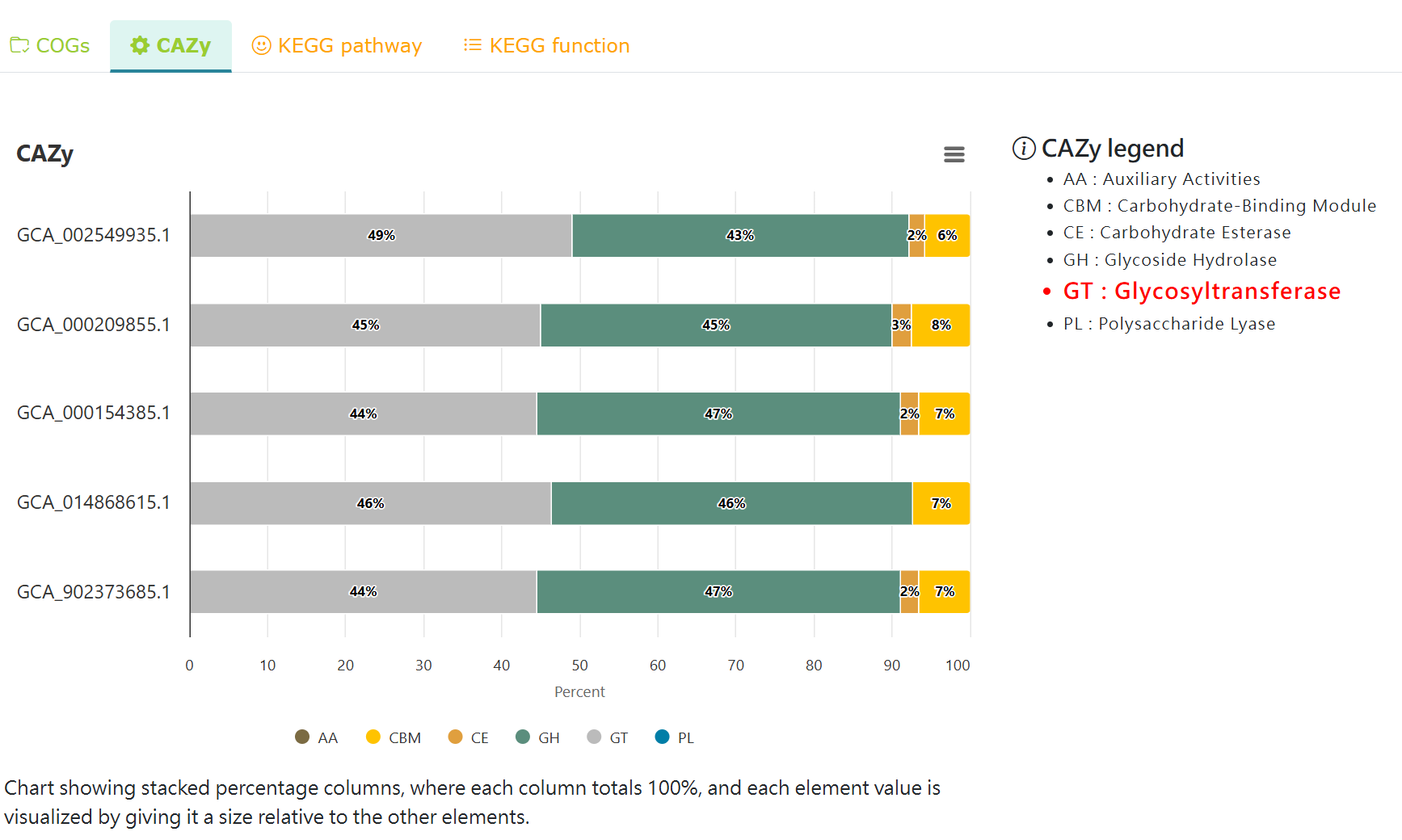
Comparative CAZy analysis of the selected strains
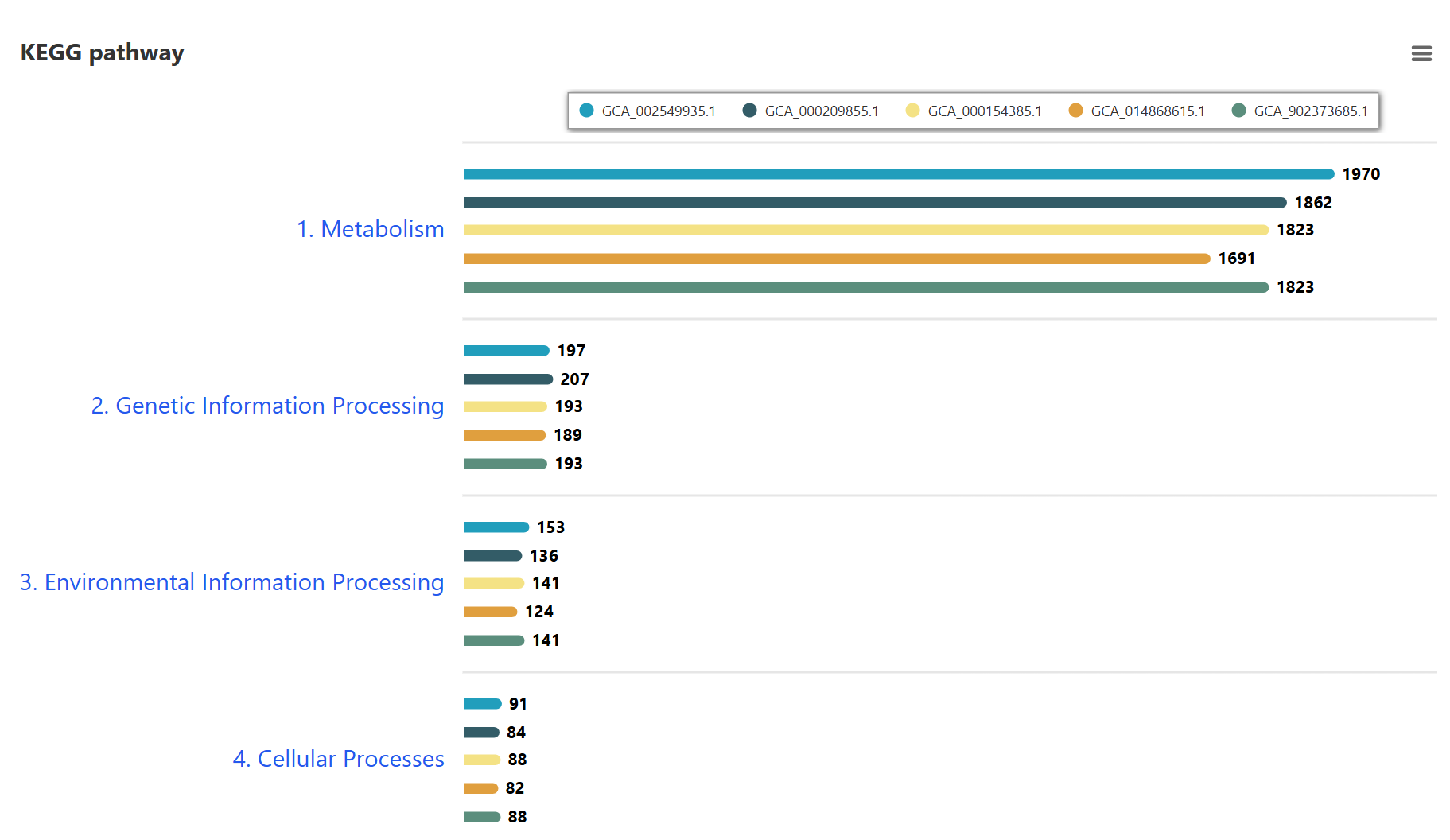
Comparative KEGG pathway analysis of the selected strains
Comparative KEGG function analysis of the selected strains
(IV)List of software and databases used in ProbioMinServer (Original vs. Updated Version)
| Function | Software | Database |
|---|---|---|
| Genome identification | ||
| Type strains comparison | Mash v2.3 | NCBI prokaryote type strains (Aug. 2023 release) |
| Average nucleotide identity (ANI) | https://github.com/chjp/ANI | - |
| Genome Circos plot | mummer2circos v1.4.2 | - |
| Circos v0.69-8 | - | |
| Safety analysis | ||
| Antibiotic resistance genes (ARGs) | RGI v6.0.0 (BLAST homolog detection) | CARD Variants v4.0.0 (July 2022 release) |
| ResFinder v4.0 | resfinder_db (May 2022 release) | |
| AMRFinderPlus v3.10.42 | (October 2022 release) | |
| Virulence factors (VFs) | BLASTN v2.8.1+ | VFDB (June 2022 release) |
| VirulenceFinder v2.0.3 | virulencefinder_db (December 2022 release) | |
| Pathogenic genes (PGs) | BLASTP v2.8.1+ | PHI-base v4.14 (November 2022 release) |
| Plasmid genes | PlasmidFinder v2.0.1 | plasmidfinder_db (January 2023 release) |
| Prophage regions | Phigaro v2.3.0 | (August 2017 release) |
| Functional analysis | ||
| Secondary metabolite gene clusters | antiSMASH v6.0.0 | (June 2022 release) |
| Primary metabolite gene clusters | gutSMASH v1.0.0 | (Oct. 2020 release) |
Updated Version – ProbioMinServer v2.0
| Function | Software | Database |
|---|---|---|
| Genome identification | ||
| Type strains comparison | Mash v2.3 | NCBI prokaryote type strains (Aug. 2023 release) |
| Average nucleotide identity (ANI) | https://github.com/chjp/ANI | - |
| Genome Circos plot | mummer2circos v1.4.2 | - |
| Circos v0.69-8 | - | |
| Genome Annotation | eggNOG-mapper v2.1.12 | eggNOG v5.0.2 (Mar. 2021 release) |
| Safety analysis | ||
| Antibiotic resistance genes (ARGs) | RGI v6.0.3 (BLAST homolog detection) | CARD Variants v4.0.2 (Nov. 2023 release) |
| ResFinder v4.6.0 | resfinder_db (Aug. 2024 release) | |
| AMRFinderPlus v4.0.3 | (Oct. 2024 release) | |
| Virulence factors (VFs) | BLASTN v2.16.0 | VFDB (Dec. 2024 release) |
| VirulenceFinder v2.0.4 | virulencefinder_db (Feb. 2024 release) | |
| Pathogenic genes (PGs) | BLASTX v2.16.0 | PHI-base v4.16 (May. 2024 release) |
| Plasmid genes | PlasmidFinder v2.1.6 | plasmidfinder_db v2.2.0 (Nov. 2024 release) |
| PlasmidHunter v1.4.5 | (May. 2024 release) | |
| Prophage regions | Phigaro v2.4.0 | (Jan. 2024 release) |
| Insertion Sequence | ISEScan v1.7.2.3 | (Apr. 2021 release) |
| Functional analysis | ||
| Secondary metabolite gene clusters | antiSMASH v7.1.0 | (Nov. 2023 release) |
| Primary metabolite gene clusters | gutSMASH v1.0.0 | (Oct. 2020 release) |
(V)Browser compatibility
| OS | Version | Chrome | Firefox | Microsoft Edge | Safari |
|---|---|---|---|---|---|
| Linux | CentOS 7 | not tested | 61.0 | n/a | n/a |
| MacOS | Big Sur, High Sierra | 96.0.4664.110 | 61.0 | n/a | 14.1.1, 13.1.2 |
| Windows | 10 | 96.0.4664.110 | 61.0 | 42.17134.1.0 | n/a |